Summary and Setup
This is a new lesson built with The Carpentries Workbench.
For this lesson, you will be working in the R programming language and the RStudio development environment on the data analysis and sharing platform CAVATICA.
Setup
Access CAVATICA
Request your free pilot fund cloud credits by contacting CAVATICA support.
For Billing group, select
Pilot Fundsif you have received free pilot fund cloud credits.Change the default setting to
Allow network access.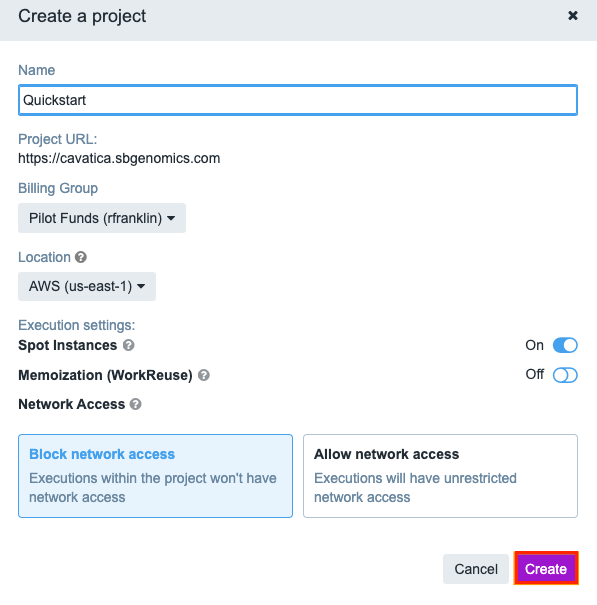
Run a Data Studio analysis. by clicking the Data Studio tab.
- Click Create new analysis.
- Select RStudio.
- Use the default Environment Setup. You can read more about default and other available environments and libraries for RStudio.
Package Installation
Next, install the required packages for this lesson. In RStudio, copy and paste the following commands into the Console:
R
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install()
# install synapser
install.packages("synapser", repos = c("http://ran.synapse.org", "https://cloud.r-project.org"))
# install tidyverse if you don't already have it
install.packages("tidyverse")
install.packages("dplyr")
install.packages("lubridate")
# load the 4 libraries you just installed
library(synapser)
library("tidyverse")
library("dplyr")
library("lubridate")
Install Bioconductor packages.
# When the following query arises, type 'a'
# Update all/some/none? [a/s/n]: a
# When the following query arises, type 'n' (it is faster)
# Do you want to install from sources the packages which need compilation?
# (Yes/no/cancel) n
BiocManager::install("clusterProfiler")
BiocManager::install("DESeq2")
BiocManager::install("AnnotationDbi")
BiocManager::install("org.Mm.eg.db")
BiocManager::install("org.Hs.eg.db")
BiocManager::install("GO.db")
BiocManager::install("EnhancedVolcano")Once the installation has finished, copy and paste the following commands into the console to verify that packages installed correctly.
library("clusterProfiler")
library("DESeq2")
library("AnnotationDbi")
library("org.Mm.eg.db")
library("org.Hs.eg.db")
library("GO.db")
library("EnhancedVolcano")Next, you will need to log in to your Synapse account.
Login option 1: Synapser takes credentials from your Synapse web session. If you are logged into the Synapse web browser, synapser will automatically use your login credentials to log you in during your R session! All you have to do is:
R
synLogin()
If for whatever reason that didn’t work, try one of these options:
Login option 2: Synapse username and password In the code
below, replace the < > with your Synapse username and
password.
R
synLogin("<username>", "<password>")
Login option 3: Synapse PAT If you usually log in to Synapse with your Google account, you will need to use a Synapser Personal Access Token (PAT) to log in with the R client. Follow these instructions to generate a personal access token, then paste the PAT into the code below. Make sure you scope your access token to allow you to View, Download, and Modify.
R
synLogin(authToken = "<paste your personal access token here>")
For more information on managing Synapse credentials with synapser, see the documentation here: https://r-docs.synapse.org/articles/manageSynapseCredentials.html.
R
# When the following query arises, type 'a'
# Update all/some/none? [a/s/n]: a
# When the following query arises, type 'n' (it is faster)
# Do you want to install from sources the packages which need compilation?
# (Yes/no/cancel) n
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("clusterProfiler")
BiocManager::install("DESeq2")
BiocManager::install("AnnotationDbi")
BiocManager::install("org.Mm.eg.db")
BiocManager::install("org.Hs.eg.db")
BiocManager::install("GO.db")
BiocManager::install("EnhancedVolcano")
Project Setup
Before you begin, determine your working directory
(get(wd) and set(wd)), and create two new
folders within your preferred working directory entitled
data and results. You can copy and paste the
following commands into the R console.
R
dir.create("data")
dir.create("results")
Data Set Download
While you can always download data from the AD Portal website via your web browser, it’s usually faster and often more convenient to download data programmatically.
Download a single file To download a single file from the AD Knowledge Portal, you can click the linked file name to go to a page in the Synapse platform where that file is stored. Using the synID on that page, you can call the synGet() function from synapser to download the file.
Exercise 1: Use Explore Data to find processed RNAseq data from the Jax.IU.Pitt_5XFAD Study.
https://adknowledgeportal.synapse.org/
This filters the table to a single file. In the “Id” column for this htseqcounts_5XFAD.txt file, there is a unique Synapse ID (synID).
We can then use that synID to download the file.
R
counts_id <- "syn22108847"
synGet(counts_id, downloadLocation = "../data/")
Bulk download files
Exercise 2: Use Explore Studies to find all metadata files from the Jax.IU.Pitt_5XFAD study.
Use the facets and search bar to look for data you want to download from the AD Knowledge Portal. Once you’ve identified the files you want, click on the download arrow icon on the top right of the Explore Data table and select “Programmatic Options” from the drop-down menu.
In the window that pops up, select the “R” tab from the top menu bar. This will display some R code that constructs a SQL query of the Synapse data table that drives the AD Knowledge Portal. This query will allow us to download only the files that meet our search criteria.
The function synTableQuery() returns a Synapse object wrapper around a CSV file that is automatically downloaded to a Synapse cache directory .synapseCache in your home directory. You can use query$filepath to see the path to the file in the Synapse cache.
R
# download the results of the filtered table query
query <- synTableQuery("SELECT * FROM syn11346063.52 WHERE ( ( `study` HAS ( 'Jax.IU.Pitt_5XFAD' ) ) AND ( `resourceType` = 'metadata' ) )")
# view the file path of the resulting csv
query$filepath
We’ll use read.csv to read the CSV file into R (although the provided read.table or any other base R version is also fine!). We can explore the download_table object and see that it contains information on all of the AD Portal data files we want to download. Some columns like the “id” and “parentId” columns contain info about where the file is in Synapse, and some columns contain AD Portal annotations for each file, like “dataType”, “specimenID”, and “assay”. This annotation table will later allow us to link downloaded files to additional metadata variables!
R
# read in the table query csv file
download_table <- read_csv(query$filepath, show_col_types = FALSE)
download_table
Finally, we use a mapping function from the purrr package to loop through the “id” column and apply the synGet() function to each file’s synID. In this case, we use purrr::walk() because it lets us call synGet() for its side effect (downloading files to a location we specify), and returns nothing.
R
# loop through the column of synIDs and download each file
purrr::walk(download_table$id, ~synGet(.x, downloadLocation = "../data/"))
An important note: for situations where you are downloading many large files, the R client performs substantially slower than the command line client or the Python client. In these cases, you can use the instructions and code snippets for the command line or Python client provided in the “Programmatic Options” menu.
